Did you know that cryo-EM has been used to uncover the molecular basis of prothrombin activation? – Investigators from St. Louis University School of Medicine and Washington University School of Medicine, in St. Louis, MO have recently solved the structure of the prothrombin-prothrombinase complex at near-atomic (4.1 Å) resolution.1 Until now, the overall structural architecture of the three macromolecular components in the prothrombin-prothrombinase complex and the molecular basis for interactions between prothrombin (fII) and the prothrombinase complex, comprising the enzyme factor Xa (fXa) and its co-factor Va (fVa), have remained elusive. This unprecedented ternary complex structure, which was obtained through cryo-electron microscopy (cryo-EM) technology, sheds great new 3-dimensional insight into prothrombin activation in blood coagulation that occurs on the surface of platelets at sites of vascular injury and to the molecular underpinnings of interaction between prothrombin and prothrombinase that triggers prothrombin cleavage and conversion to thrombin. This significant leap forward in understanding may lead to new therapeutic treatments for thrombotic and hemophilic diseases.
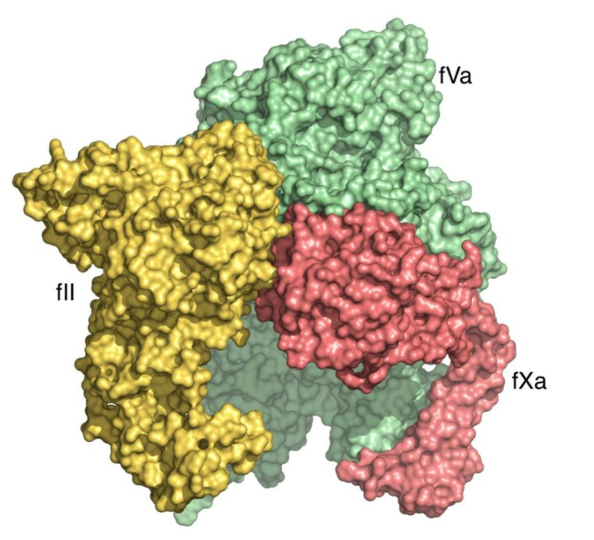
The prothrombin-fVa-fXa complex is shaped like a macromolecular dome with a domain of fVa at the top (the A2 domain) and a bottom base, comprised of domains of prothrombin, fVa and fXa, in optimal alignment for membrane binding (such as on the membranes of human platelets or red blood cells). The complex structure provides molecular evidence that the function of cofactor fVa is to enhance prothrombin activation by providing a scaffold for assembly of the prothrombin-fXa complex rather than allosterically changing the conformation of fXa. These unprecedented details of molecular interaction illustrate for the first time, how factors Va and Xa associate on the membrane surface, as well as how the inactive (zymogen) form of prothrombin binds in a conformation that leads to the generation of its activated form, thrombin, which is central to hemostasis. The presence of fVa in the prothrombinase complex significantly increases the rate of prothrombin conversion to thrombin and this new complex structure reveals how the A2 domain of the fVa cofactor orchestrates prothrombin activation along 1 of 2 possible pathways, the meizothrombin pathway, following initial cleavage at arginine 320. Elucidation of the molecular basis of the prothrombin-prothrombinase interaction has great significance to the large class of trypsin-like zymogens which prothrombin represents and provides a potential doorway into solving the structures of more membrane-associated coagulation complexes in the future.
Learn more here.
